Arnau Sebé-Pedrós
Group leader CRG; Associate Faculty ToL Sanger Institute.
Genome regulation, chromatin, cell types, and evolution. https://www.sebepedroslab.org
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-PedrósDelighted to see our work now published in @currentbiology.bsky.social. A great place for Bostrychia to stake its claim as an emerging red algal model system. Check it out here: www.cell.com/current-biol...
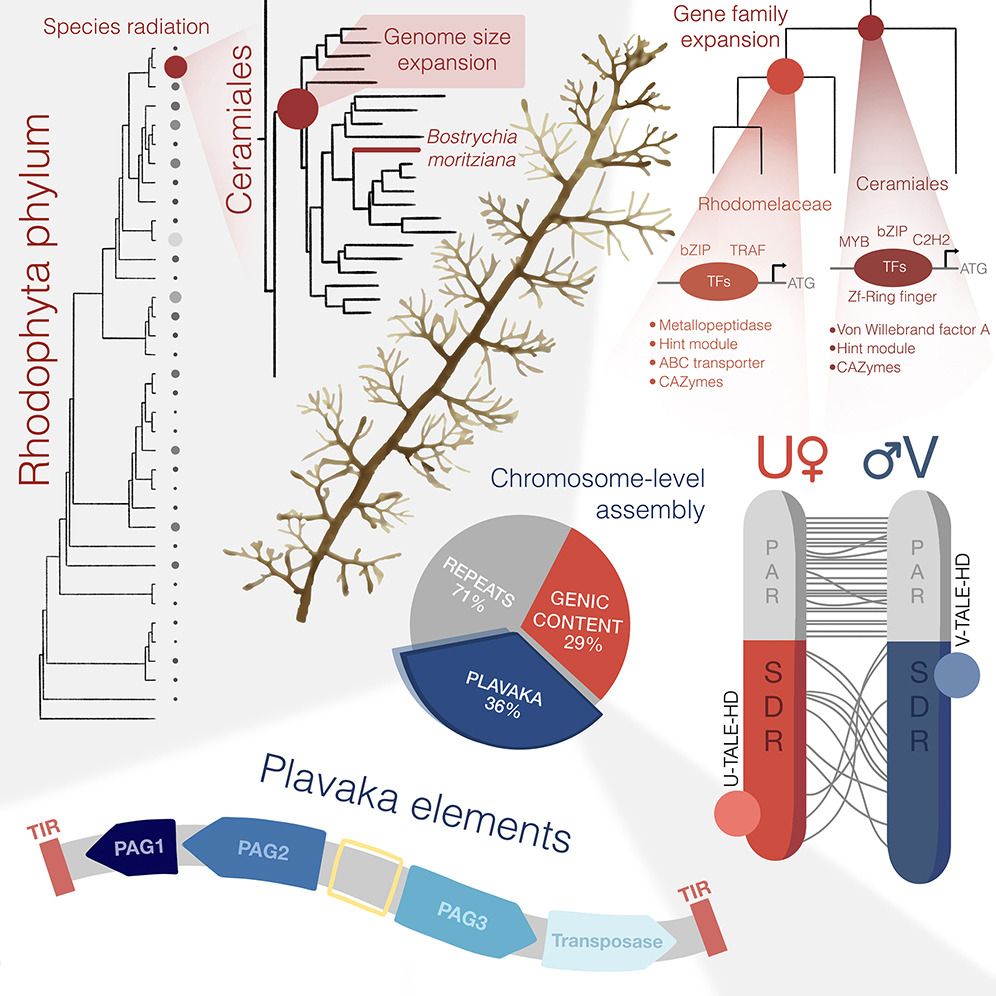
- 🚨Preprint Alert🚨 I'm really happy to finally share our exploits in red algae and introduce the wider community to Bostrychia, the new model system we have been developing to tackle the molecular biology of red algae. Isn't she a beauty? Thread below 🧵 1/8 www.biorxiv.org/content/10.1...
- Reposted by Arnau Sebé-PedrósOut in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis 🧬🩸 screen of fully synthetic enhancers in blood progenitors 🤖 AI that creates new cell state specific enhancers 🔍 negative synergies between TFs lead to specificity! www.cell.com/cell/fulltex... 🧵
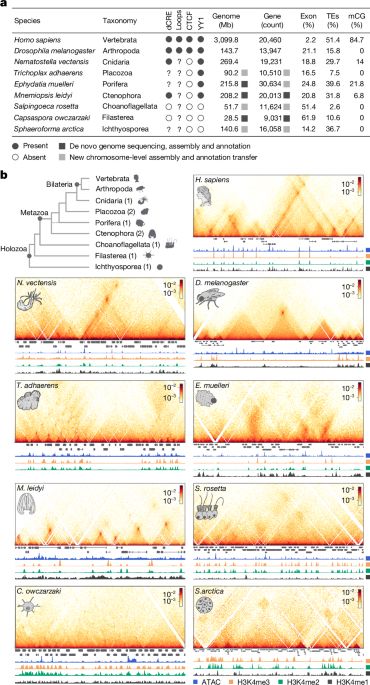
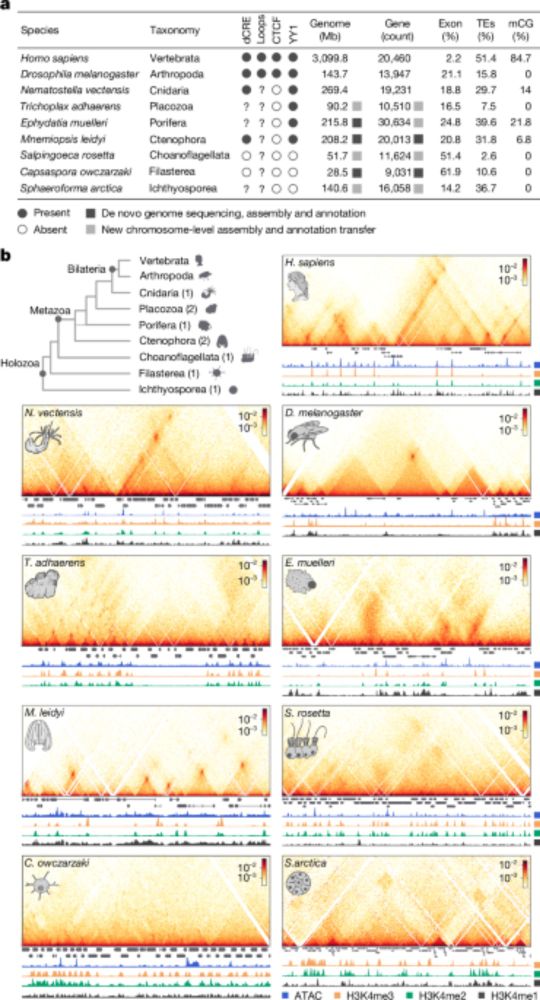
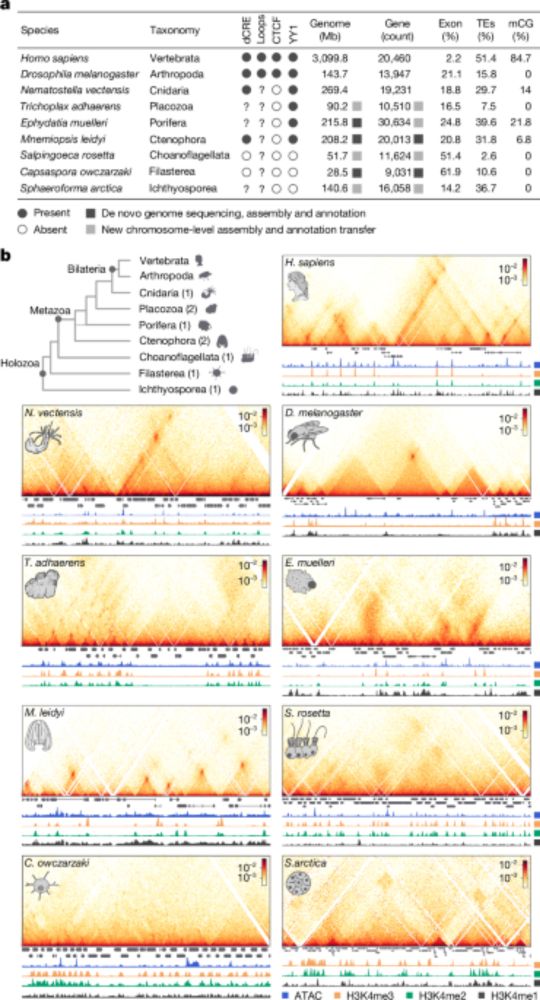
- Reposted by Arnau Sebé-PedrósChromatin loops and the origin of animals www.nature.com/articles/s41...
- Check out our latest work on the evolution of animal genome regulation out today in @nature.com. Nicely summarized below by @ianakim.bsky.social. www.nature.com/articles/s41... This is a major output from our ERC-StG project Evocellmap @erc.europa.eu at @crg.eu
- I’m very excited to share our work on the early evolution of animal regulatory genome architecture - the main project of my postdoc, carried out across two wonderful and inspirational labs of @arnausebe.bsky.social and @mamartirenom.bsky.social. www.nature.com/articles/s41...
- Reposted by Arnau Sebé-PedrósCome and join us! We’re hiring a new Group Leader in Generative Biology at the @sangerinstitute.bsky.social Building AI models or the data to train them? Core funding of >$130M a year for a faculty of ~30. www.nature.com/naturecareer... acrobat.adobe.com/id/urn:aaid:... pls RT!
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-PedrósIt's publication day of my first book: The Tree of Life. The tree of life is a time machine that can take us back 4 billion years to meet our most distant ancestor. It is the magic that lets us tell the origin stories, beginning with this ancient relative, of everything from mushrooms to man.
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós#multicellularity lovers: the website for the EMBO workshop in multicellularity is live. Barcelona is waiting for you! #protistsonsky
- Reposted by Arnau Sebé-PedrósPreprint on hifiasm Nanopore-only assembly. Led by Haoyu Cheng: www.biorxiv.org/content/10.1...
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
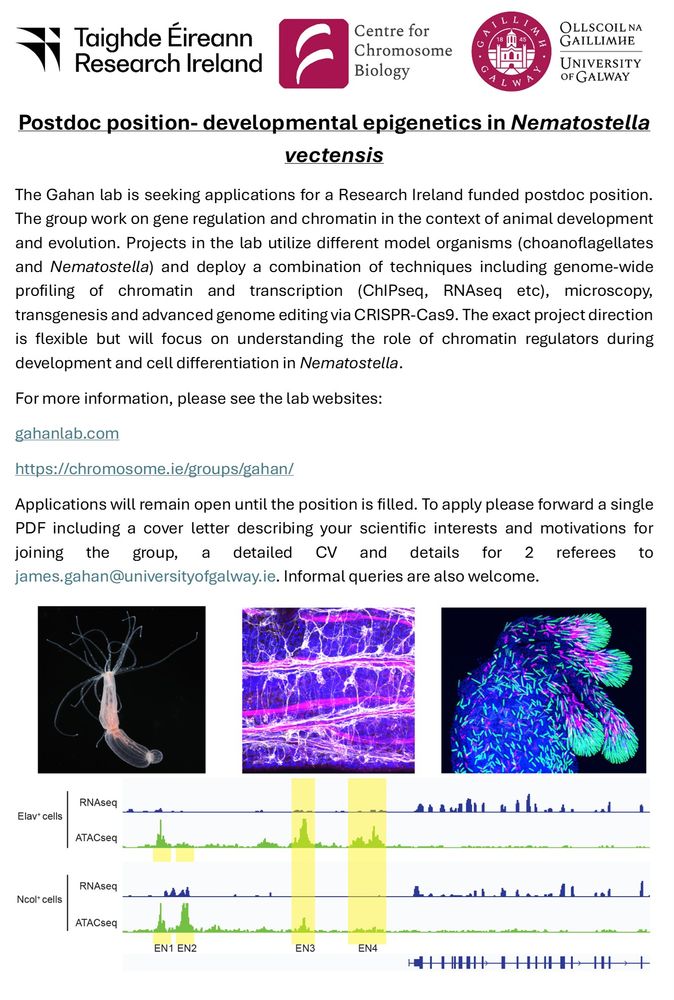
- Reposted by Arnau Sebé-PedrósMy group is hiring a new Postdoc to work on gene regulation and chromatin in Nematostella. Please RT! For more information see here: gahanlab.com tinyurl.com/34xke35d
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-PedrósExcited to see all the EvoChromo people again at this meeting! A fantastic collection of kind and curious researchers made the last edition the top meeting I've attended, and I expect v2 to be even better!
- Registrations are now open for the next @embo.org EvoChromo meeting in December. meetings.embo.org/event/24-evo... Join us in beautiful Costa Brava to discuss chromatin evolution! Co-organized with @tobiaswarnecke.bsky.social @sandraduharcourt.bsky.social @levine-lab.bsky.social and Nick Irwin
- Registrations are now open for the next @embo.org EvoChromo meeting in December. meetings.embo.org/event/24-evo... Join us in beautiful Costa Brava to discuss chromatin evolution! Co-organized with @tobiaswarnecke.bsky.social @sandraduharcourt.bsky.social @levine-lab.bsky.social and Nick Irwin
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-PedrósVery proud of two new preprints from the lab: 1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...
- Reposted by Arnau Sebé-PedrósWe released our preprint on the CREsted package. CREsted allows for complete modeling of cell type-specific enhancer codes from scATAC-seq data. We demonstrate CREsted’s robust functionality in various species and tissues, and in vivo validate our findings: www.biorxiv.org/content/10.1...
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Check out our latest work on chromatin evolution by @crisnava.bsky.social and @seanamontgomery.bsky.social. Building on our study of histone mark conservation (www.nature.com/articles/s41...), we now explore whether these same marks define similar chromatin states across eukaryotes.
- Excited to share this story from the “Chromatin Dream Team” in the @arnausebe.bsky.social lab on chromatin evo across eukaryotes! 12 histone mark profiles from 12 species, including rhizarians, discobans, and cryptomonads. Read on to see what we found! www.biorxiv.org/content/10.1... 1/n
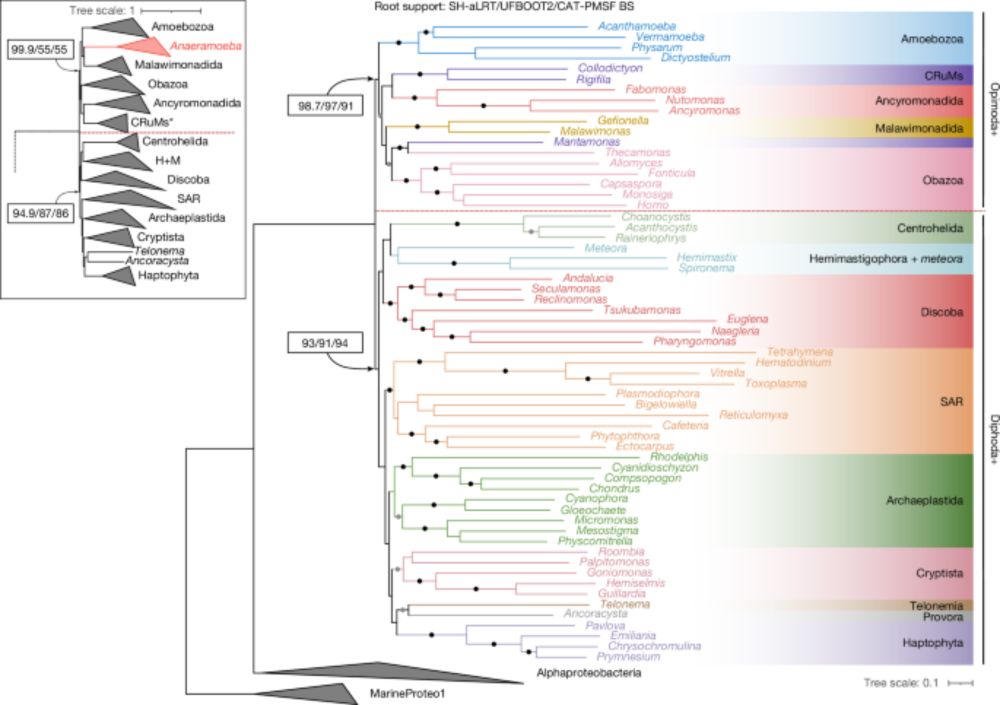
- Reposted by Arnau Sebé-Pedrós🚨 New paper alert! 🚨 A robustly rooted tree of eukaryotes sheds light on their deep evolutionary ancestry—suggesting that the Last Eukaryotic Common Ancestor (LECA) may have had an excavate-like cell architecture. 🧵🔬 (1/) www.nature.com/articles/s41...
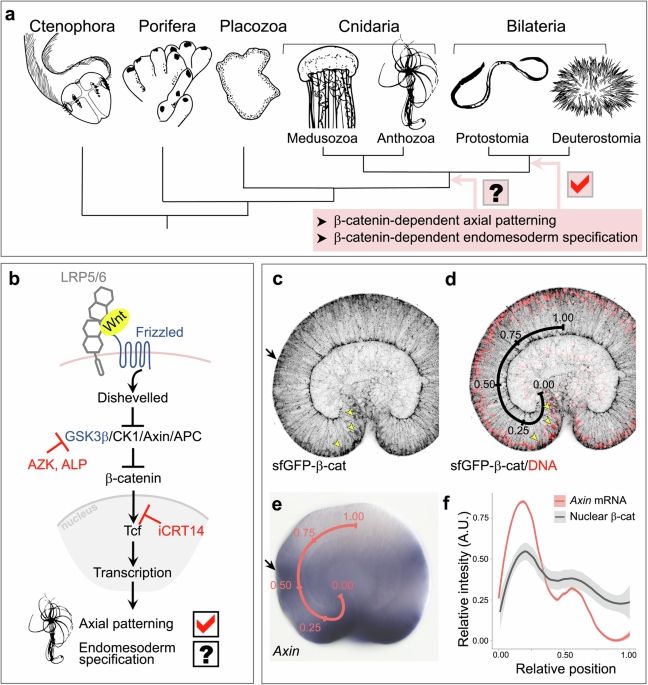
- Reposted by Arnau Sebé-PedrósTanya's @tclebedeva.bsky.social paper on endomesoderm specification in the beta-catenin-negative area of the Nematostella embryo is out! www.nature.com/articles/s41...
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
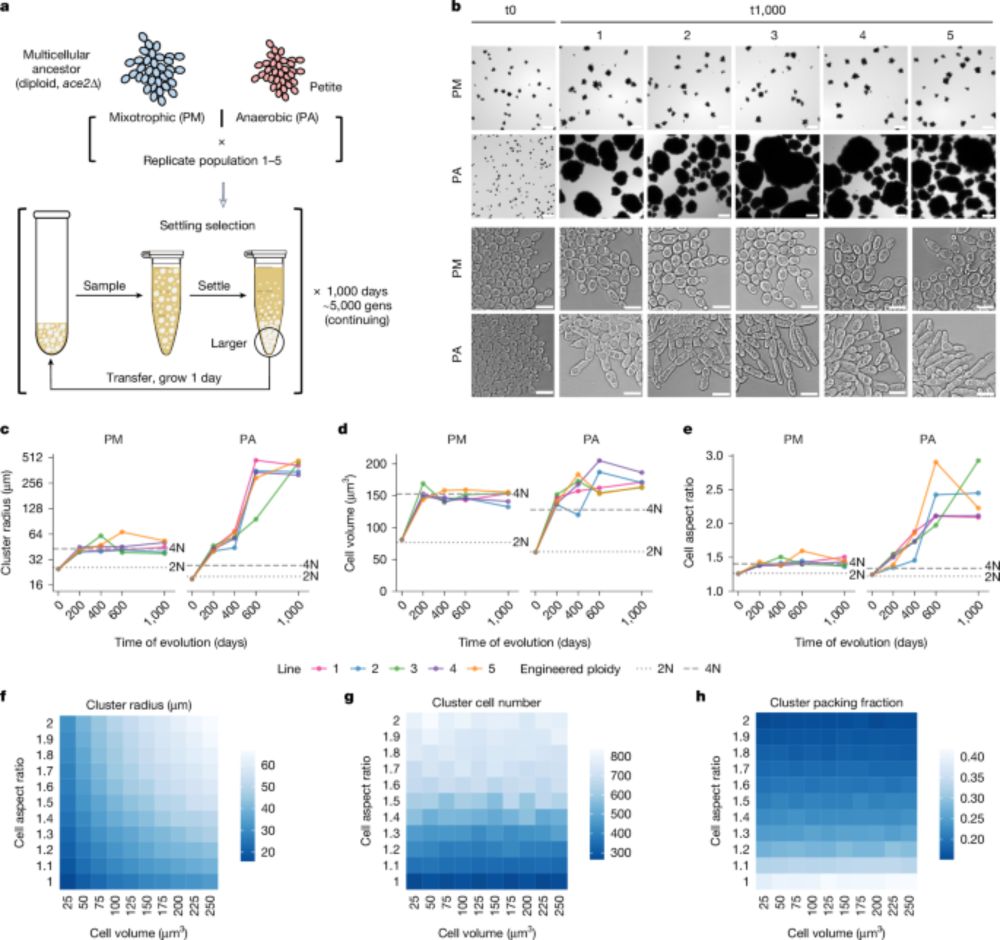
- Reposted by Arnau Sebé-Pedrós1/46 Hey folks, we have a new paper out on the MuLTEE. Strap in and I’ll tell you the story of how this “little paper on polyploidy” turned into the most data rich paper our lab has produced, largely thanks to the leadership and work ethic of @kaitong25.bsky.social. www.nature.com/articles/s41...
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- We offer three new positions to join BCA Phase 0: 1. A full-stack developer to work on the BCA database with @ebi.embl.org 2. A senior research technician to develop sc methods 3. A bioinformatician to analyze new atlases with @sangerinstitute.bsky.social Details below. Please, share and repost.
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- One week left to apply!
- Reposted by Arnau Sebé-PedrósDo you use Darwin Tree of Life (DToL) genomes for your work or grant applications? If so, can you please fill out this form? docs.google.com/forms/d/e/1F... We are gathering info on how DToL genomes are used. It will help us raise money to continue producing publicly available high-quality genomes.
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-PedrósThis has been a fantastic adventure - to capture the genomic regulatory code underlying brain cell types (using deep learning models trained on chromatin accessibility), and then use these models to compare cell types between the bird and mammalian brain
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós🚨Preprint Alert🚨 I'm really happy to finally share our exploits in red algae and introduce the wider community to Bostrychia, the new model system we have been developing to tackle the molecular biology of red algae. Isn't she a beauty? Thread below 🧵 1/8 www.biorxiv.org/content/10.1...
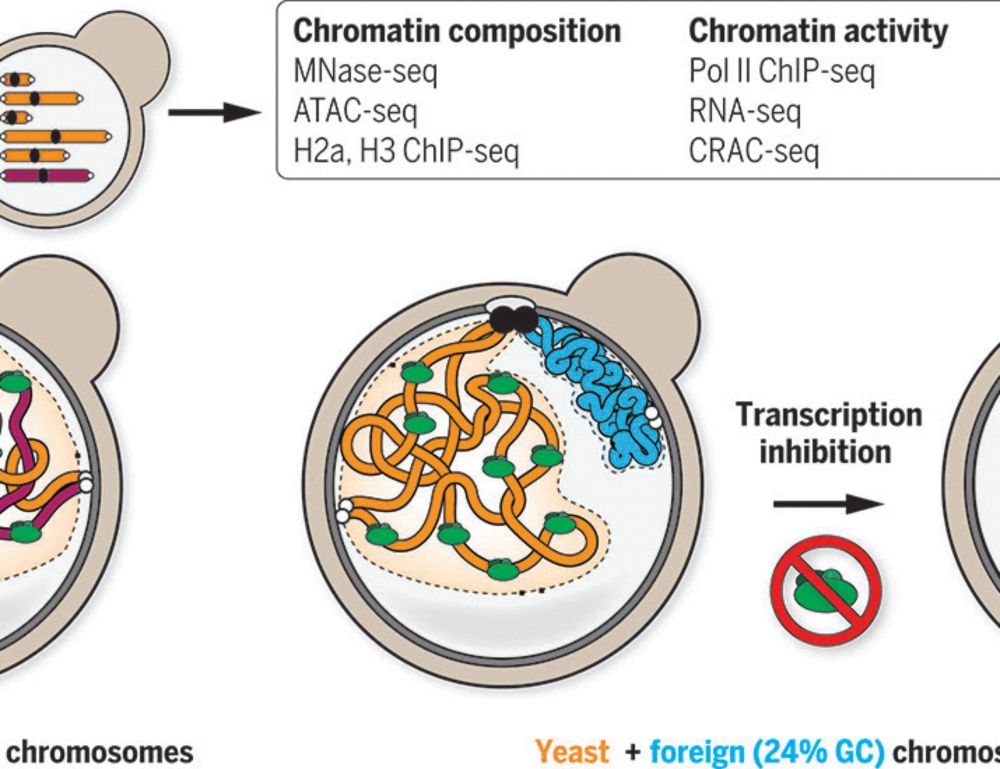
- Reposted by Arnau Sebé-PedrósOur latest work: how can compartmentalization emerge in a eukaryotic genome lacking canonical heterochromatin? By investigating bacterial genomes put in yeast, we show that the presence or absence of transcription is sufficient! #chromatin #3Dgenome #generegulation www.science.org/doi/10.1126/... 👇
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- One month left to apply!
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]
- Reposted by Arnau Sebé-Pedrós[Not loaded yet]