Sean Montgomery
Postdoc in the Sebé-Pedrós lab at the CRG studying heterochromatin diversity and function across eukaryotes. Can be found chasing frisbees when not in the lab.
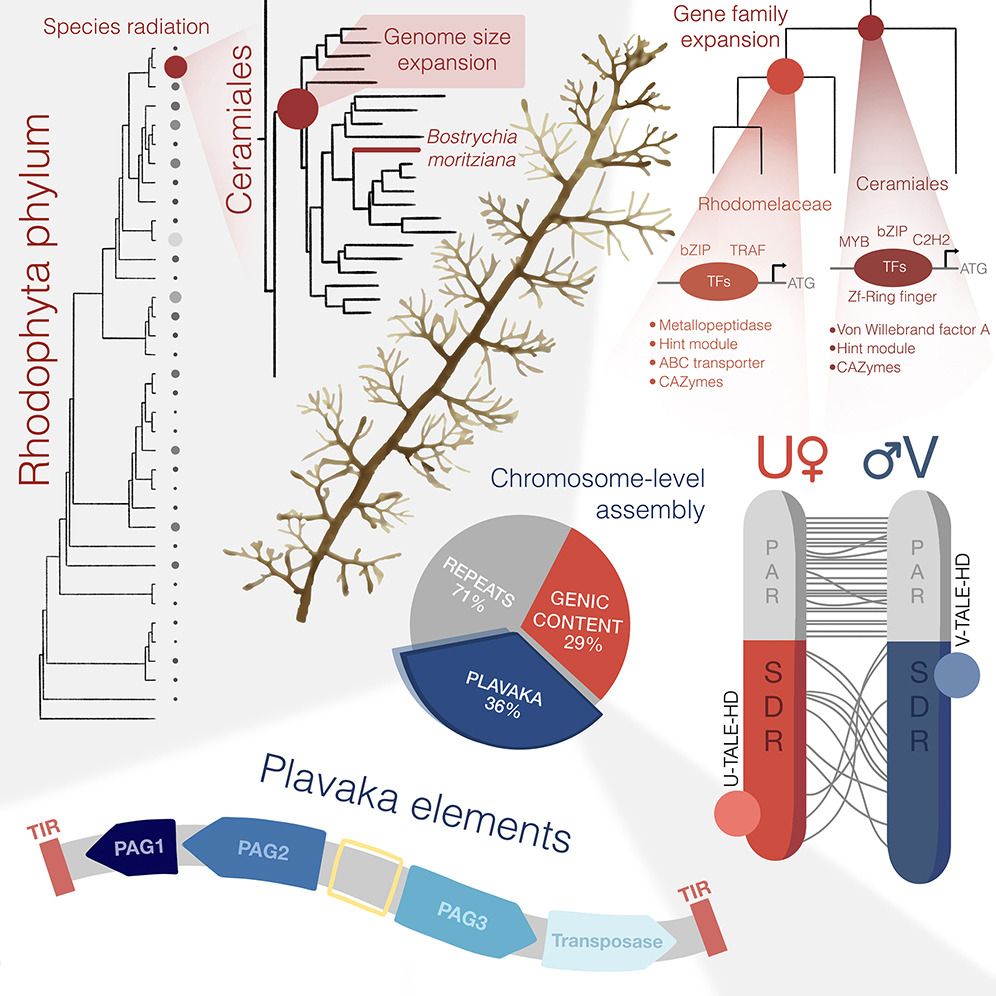
- Reposted by Sean MontgomeryDelighted to see our work now published in @currentbiology.bsky.social. A great place for Bostrychia to stake its claim as an emerging red algal model system. Check it out here: www.cell.com/current-biol...
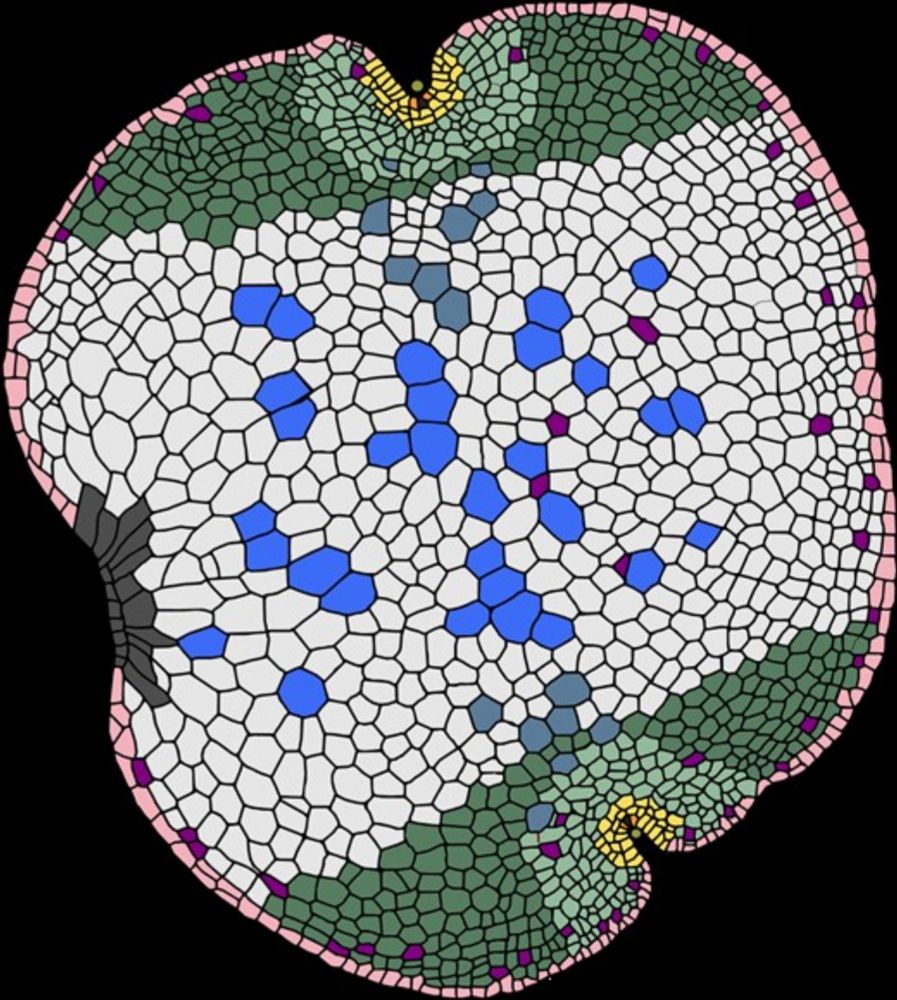
- 🚨Preprint Alert🚨 I'm really happy to finally share our exploits in red algae and introduce the wider community to Bostrychia, the new model system we have been developing to tackle the molecular biology of red algae. Isn't she a beauty? Thread below 🧵 1/8 www.biorxiv.org/content/10.1...
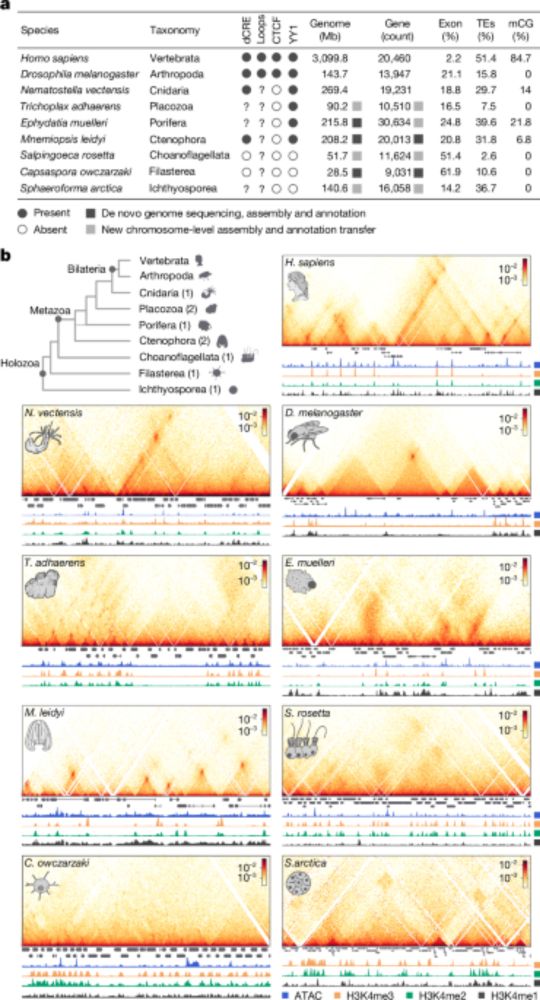
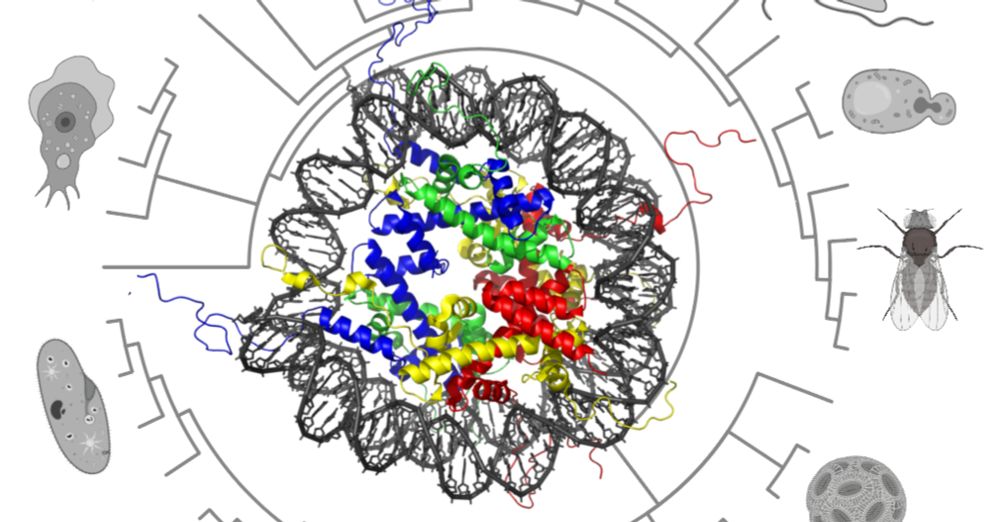
- Check out this tremendous work from @ianakim.bsky.social! An enormous effort to gather micro-C data from 9 species and uncover insights into how animal genome organization evolved and diversified!
- I’m very excited to share our work on the early evolution of animal regulatory genome architecture - the main project of my postdoc, carried out across two wonderful and inspirational labs of @arnausebe.bsky.social and @mamartirenom.bsky.social. www.nature.com/articles/s41...
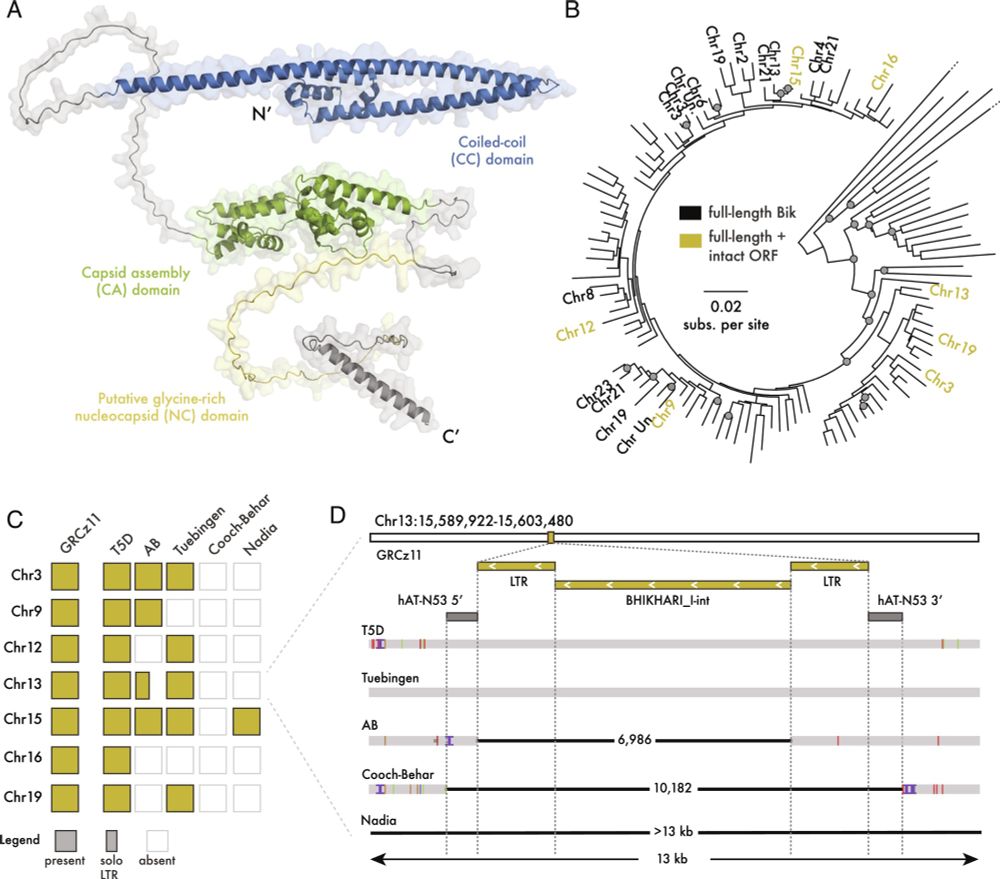
- Reposted by Sean Montgomery💥🥳 At long last, our latest paper is out! Gag proteins of endogenous retroviruses are required for zebrafish development www.pnas.org/doi/10.1073/... Led heroically by Sylvia Chang & @jonowells.bsky.social A study which has changed the way I think of #transposons! No less! 🧵 1/n
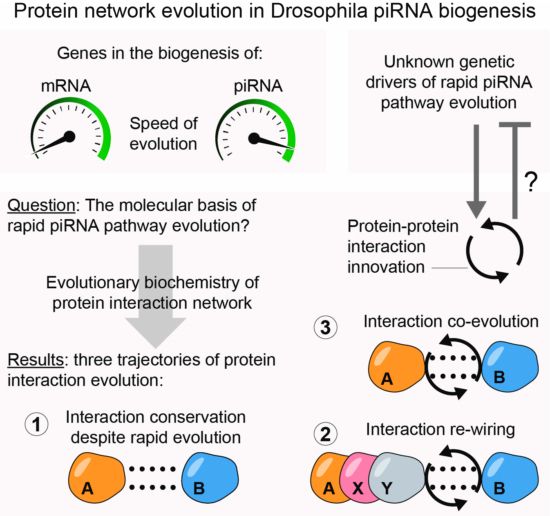
- Reposted by Sean Montgomery🧵 reprise: our paper on protein network evolution in the piRNA genome defence pathway has now been published at @embojournal.org 🥳 Very positive experience with fast and constructive reviewing (thank you, three secret people) and good journal communication. bsky.app/profile/embo...
- Comprehensive mapping of protein-protein interactions in the Drosophila piRNA pathway reveals how adaptive evolution contributes to rapid diversification of this genome-defense mechanism Peter Andersen @germline.bsky.social and collaborators www.embopress.org/doi/full/10....
- Reposted by Sean MontgomeryOur preprint has now found a wonderful home in @molbioevol.bsky.social. Shout out to our collaborators in the Mozgova lab too 💚
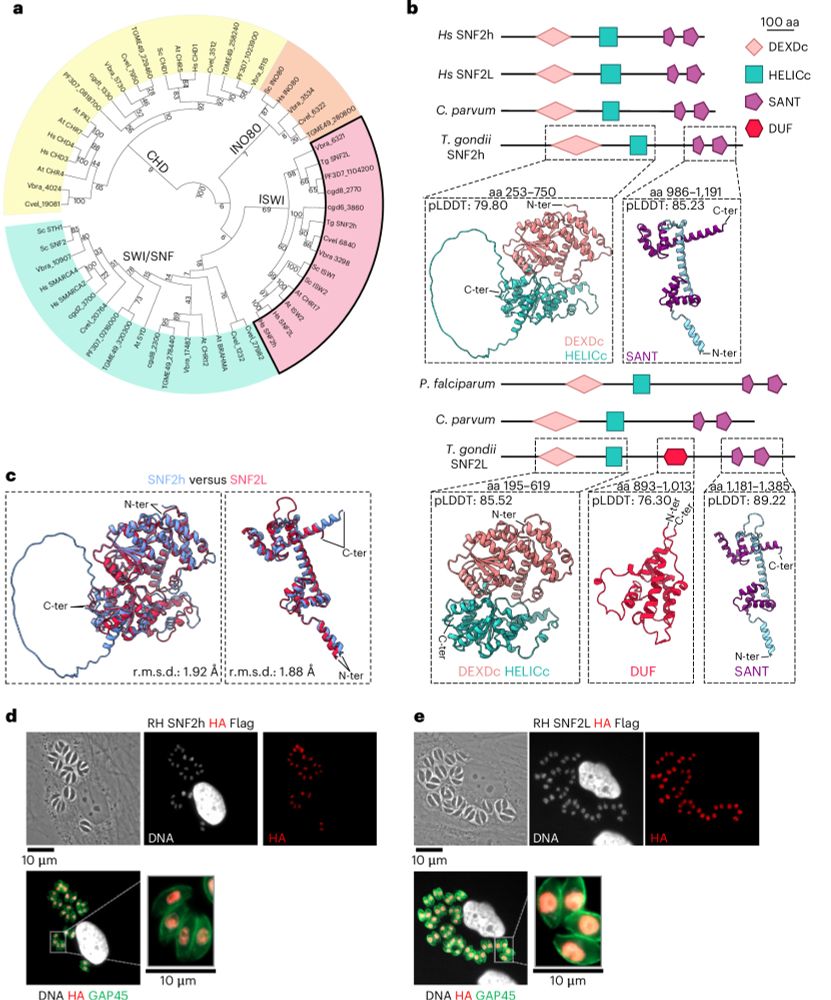
- Reposted by Sean MontgomeryIn the silence of chromatin, Toxoplasma whispers change. TgSNF2h sculpts the genome - MORC in the backseat. Some genes wake. Others sleep. A dance of nucleosomes shaping space for life parasite stages to unfold. Now in Nature Microbiology: rdcu.be/ehkWp
- Excited to see all the EvoChromo people again at this meeting! A fantastic collection of kind and curious researchers made the last edition the top meeting I've attended, and I expect v2 to be even better!
- Registrations are now open for the next @embo.org EvoChromo meeting in December. meetings.embo.org/event/24-evo... Join us in beautiful Costa Brava to discuss chromatin evolution! Co-organized with @tobiaswarnecke.bsky.social @sandraduharcourt.bsky.social @levine-lab.bsky.social and Nick Irwin
- Fantastic study on so many levels, check it out! DNA methylation, chromatin accessibility, developmental biology, horizontal gene transfer, and my favourite, Marchantia! This paper has it all
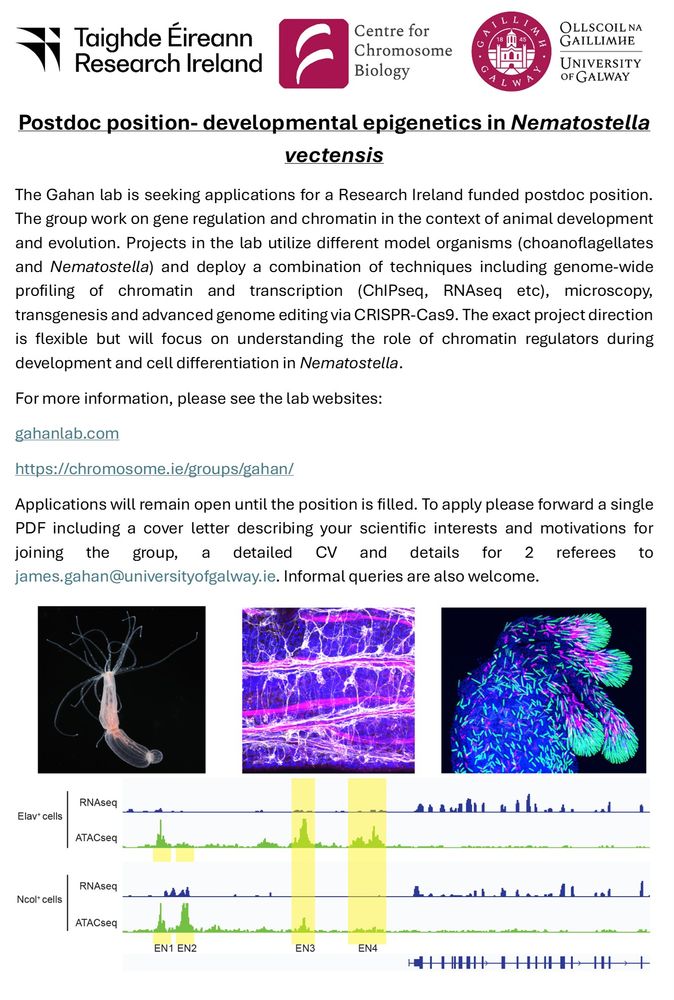
- Reposted by Sean MontgomeryMy group is hiring a new Postdoc to work on gene regulation and chromatin in Nematostella. Please RT! For more information see here: gahanlab.com tinyurl.com/34xke35d
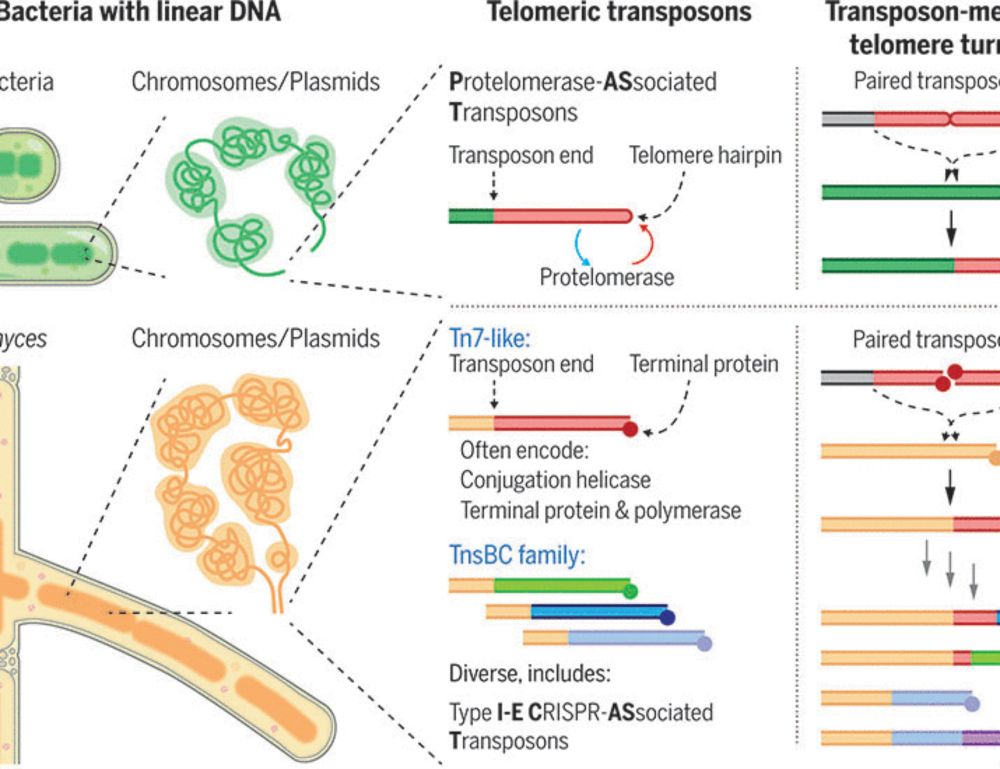
- Reposted by Sean Montgomerywow, telomeric transposons in bacteria with linear chromosomes! (of course this was first figured out in flies, inc by Bob Levis, who i was happy to see few days ago at the fly meeting). 🪰 www.science.org/doi/10.1126/... www.sciencedirect.com/science/arti... www.sciencedirect.com/science/arti...
- Reposted by Sean MontgomeryInterested in photosynthetic organisms? Looking for a space to focus on research and kick-start your journey as a group leader? We’d love for you to join us. We offer an incredibly competitive package—on par with about 2 ERC grants for 8 years—and a collaborative environment unlike anywhere else 1/2
- Reposted by Sean MontgomeryRelaxed DNA substrate specificity of transposases involved in programmed genome rearrangement 🤯 #TEsky www.biorxiv.org/content/10.1...
- Reposted by Sean Montgomery[Not loaded yet]
- Excited to share this story from the “Chromatin Dream Team” in the @arnausebe.bsky.social lab on chromatin evo across eukaryotes! 12 histone mark profiles from 12 species, including rhizarians, discobans, and cryptomonads. Read on to see what we found! www.biorxiv.org/content/10.1... 1/n
- Reposted by Sean Montgomery1/ Transposable elements are often called "jumping genes" because they mobilize within genomes. 🧬 But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯 Our new study reveals how retrotransposons invade the germline directly from somatic cells. www.biorxiv.org/content/10.1... A short thread 🧵👇
- Reposted by Sean Montgomery1/ H3K27me3 mimicry has repeatedly emerged through evolution, but what's the physiological relevance? We show that JARID2 and PALI1 mimic H3K27me3 to antagonise PRC2 in vivo and restrict the spread of Polycomb domains. 🧵 www.biorxiv.org/content/10.1...
- Reposted by Sean Montgomery[Not loaded yet]
- Reposted by Sean Montgomery[Not loaded yet]
- Reposted by Sean Montgomery🚨Preprint Alert🚨 I'm really happy to finally share our exploits in red algae and introduce the wider community to Bostrychia, the new model system we have been developing to tackle the molecular biology of red algae. Isn't she a beauty? Thread below 🧵 1/8 www.biorxiv.org/content/10.1...
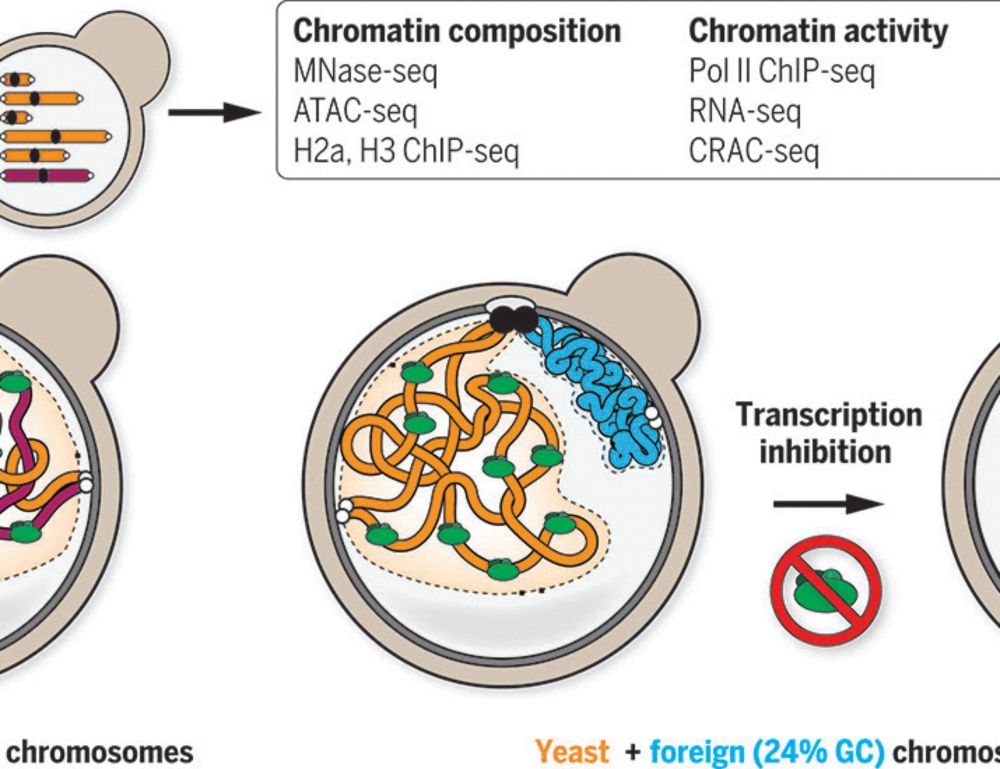
- Reposted by Sean MontgomeryOur latest work: how can compartmentalization emerge in a eukaryotic genome lacking canonical heterochromatin? By investigating bacterial genomes put in yeast, we show that the presence or absence of transcription is sufficient! #chromatin #3Dgenome #generegulation www.science.org/doi/10.1126/... 👇
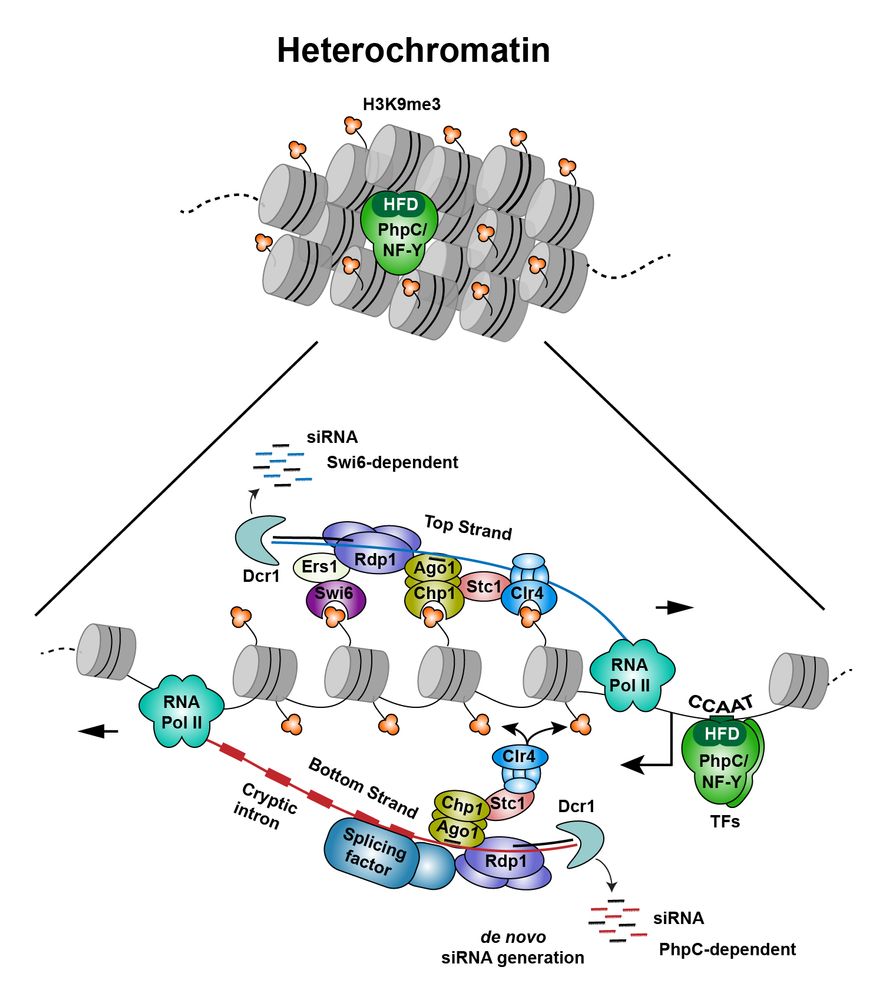
- Reposted by Sean Montgomery🔬Decoding Silencing: How Heterochromatin Transcribes to Silence Itself! 🧬 Discover how a pioneer transcription factor-like complex infiltrates repressive heterochromatin, producing transcripts with hidden introns that kickstart RNAi-mediated heterochromatin formation www.nature.com/articles/s41...
- Intriguing work on an H3K27me3-reader protein leading to transcriptional activation: www.embopress.org/doi/full/10.... Ciliates continue to impress! The authors also draw nice parallels to Rhino (H3K9me3 reader) and piRNA cluster expression in Drosophila
- Reposted by Sean MontgomeryBy definition, enhancers can activate from a distance. But with increased distance between enhancer and promoter, the activation drops. To study this systematically, we build a synthetic locus: www.cell.com/molecular-ce... 1/12
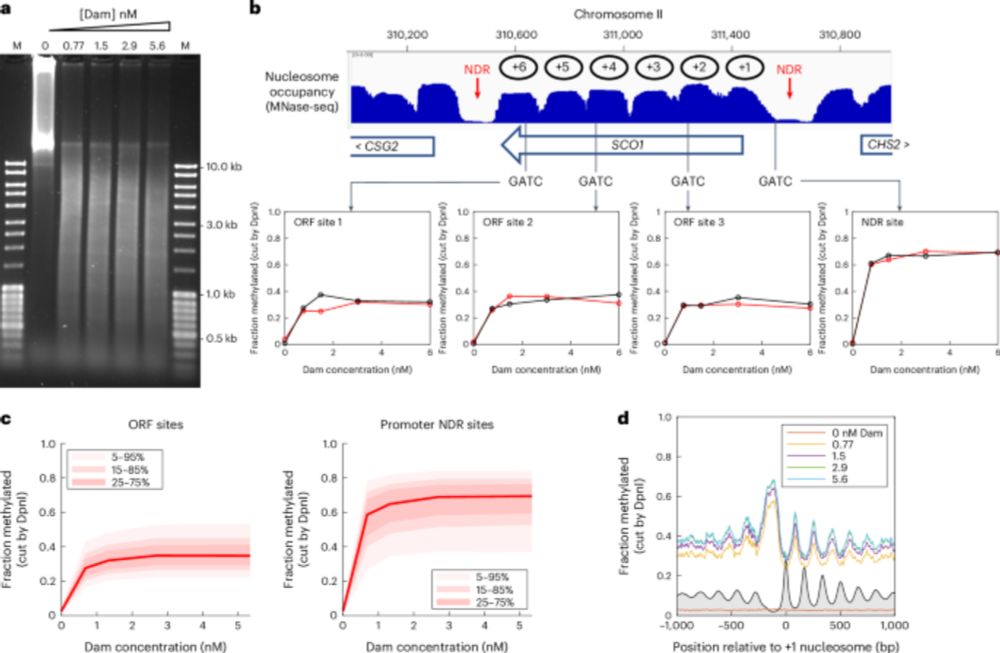
- Reposted by Sean MontgomeryThe yeast genome is globally accessible in living cells [new from the group of David Clark] www.nature.com/articles/s41... ▶️Measured DNA accessibility in living budding yeast by inducible expression of methyltransferases ▶️The genome is globally accessible in living cells, unlike in isolated nuclei
- Reposted by Sean Montgomerybehold our paper on Medusavirus nucleosomes. Fantastic work by Chelsea Toner, @nhoitsma.bsky.social, and Sashi Weerawarana This giant virus also encodes a linker histone, more about that in the paper 🧪👩🔬 🧵. www.nature.com/articles/s41...
- Reposted by Sean Montgomery[Not loaded yet]
- Check out the lab website for more details on what we do! www.sebepedroslab.org
- Reposted by Sean Montgomery[Not loaded yet]
- Very nice to see this work on silencers from the Stark lab! doi.org/10.1016/j.mo...
- Reposted by Sean Montgomery[Not loaded yet]
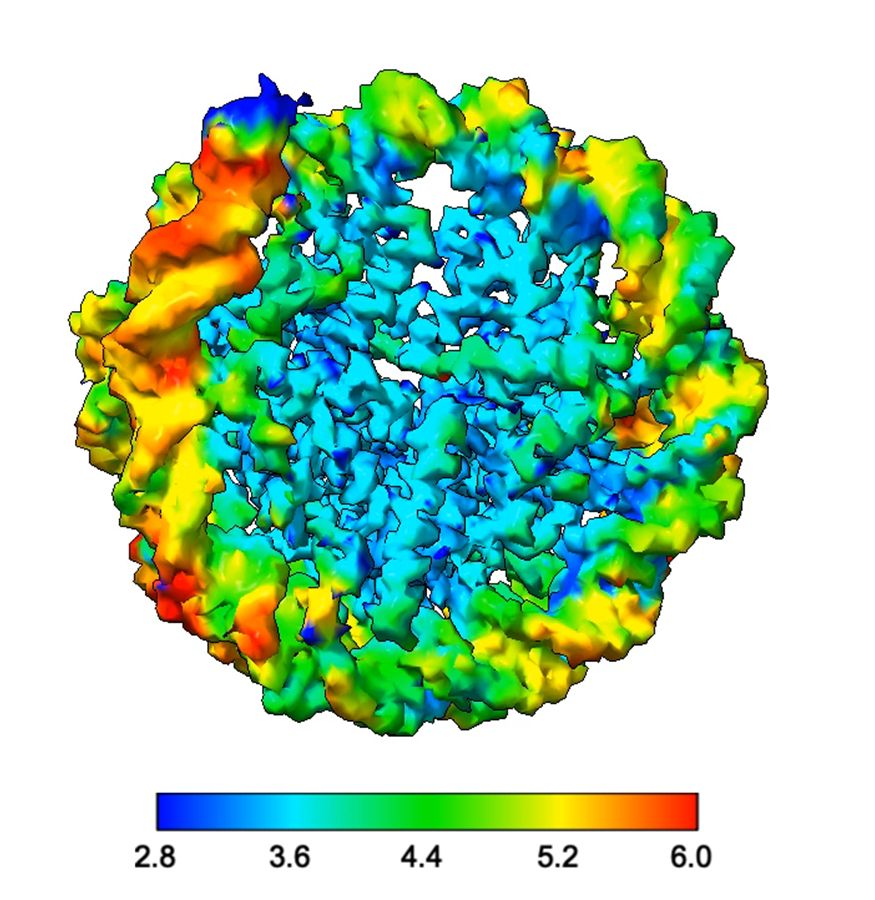
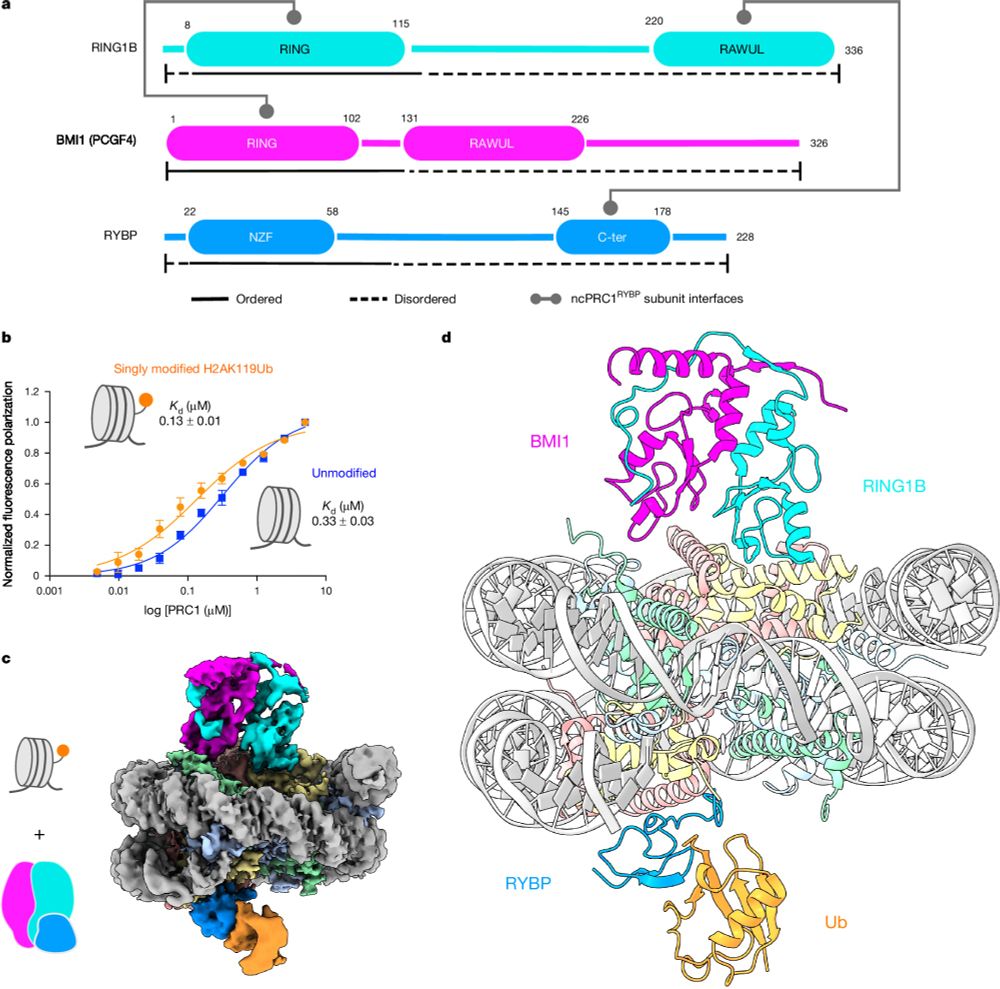
- Reposted by Sean MontgomeryPlease see our paper in Nature on read-write mechanisms of H2AK119 ubiquitination by Polycomb repressive complex I. Congrats to the whole team, especially Victoria and huge thanks to our collaborator JP Armache! Also big thanks to Mark Foundation for Cancer Research for the support! rdcu.be/dZ5HZ
- Reposted by Sean MontgomeryInterested in chromatin architecture? Checkout our new method - PCP - that can map genome organization as multi-way interactions as well as nucleosome footprint and nucleosome spacing at very high resolution. www.biorxiv.org/content/10.1...
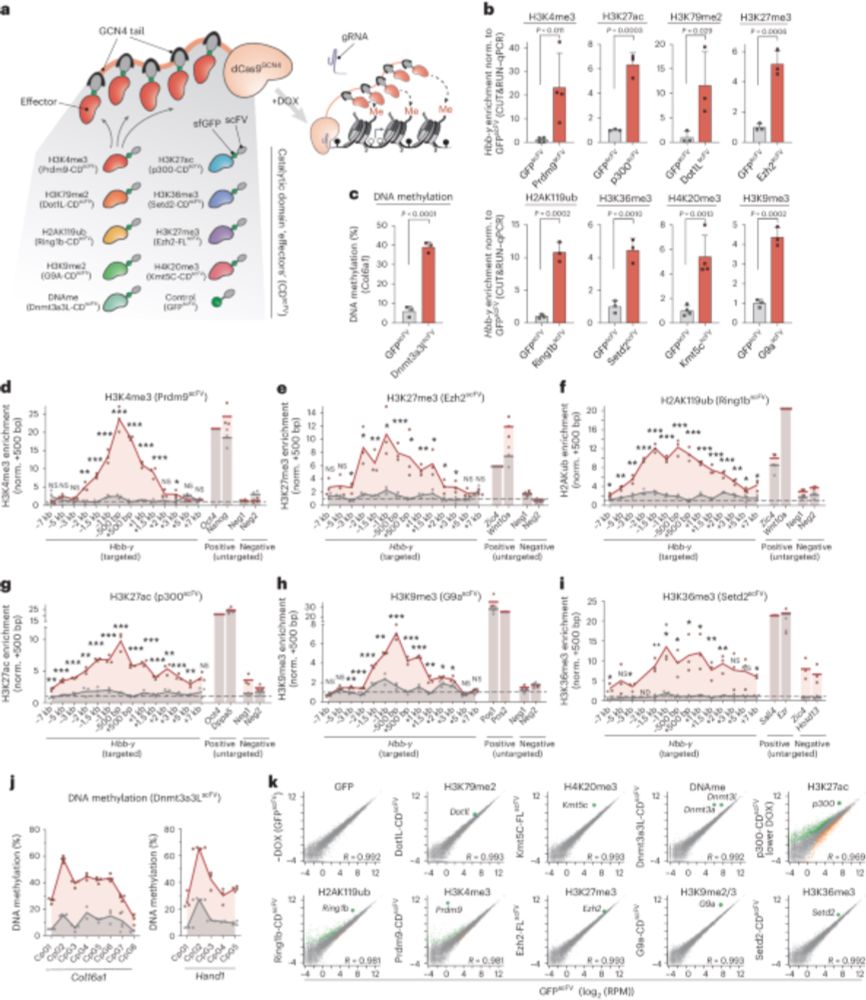
- Reposted by Sean MontgomeryI've been meaning to write a lengthy thread about our experiences with DNA methylation editing in mouse ESCs if that would be useful (or interesting?) for a niche audience here. tl;dr: it was frustrating but we learned a lot, and it ended with our shiny new pub (1/17) www.nature.com/articles/s41...
- Reposted by Sean MontgomeryOur lab is seeking a MSc student to work on the developmental dynamics of chromatin in the sea anemone Nematostella vectensis. We're looking for a student with both dry and wet lab expertise in genomics/bioinformatics, and intetest in development and epigenetics.
- Reposted by Sean Montgomery[Not loaded yet]
- Reposted by Sean Montgomery📜 An atlas of conserved transcription factor binding sites reveals the cell type- resolved gene regulatory landscape of flowering plants 🧑🔬 Leo A. Baumgart, Abraham Morales-Cruz, Sharon I. Greenblum, Ronan C O'Malley,et al. 📔 bioRxiv 🔗 www.biorxiv.org/content/10.1... #️⃣ #PlantScience #PlantEvoDevo
- Reposted by Sean MontgomeryIntriguing findings by Angelique Deleris & co: DNAme in plants is required for H3K27me3 deposition at new TE insertions 👀 This is surprising as those marks were previously thought to be competing! www.biorxiv.org/content/10.1...
- Reposted by Sean MontgomeryBacterium: “Archaea? Yeah, they’re alright. Quiet, peace-lovi....aaaargggh, STOP! STOP IT! WHAT IS HAPPENING??” [silence] Archaeon: “Mwahahaha” (probably) Discover the dark side of archaea in our latest pre-print, led by Romain Strock biorxiv.org/content/10.1... 1/10
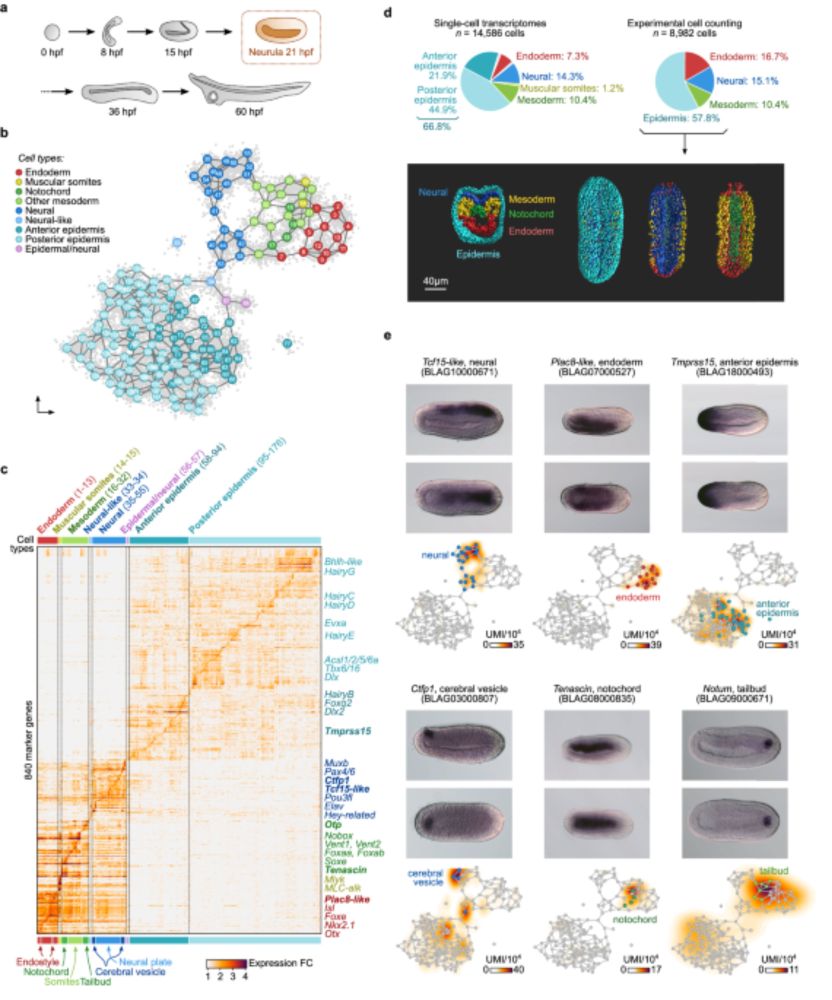
- Reposted by Sean MontgomeryOur work on the evolution of head mesoderm development in chordates is finally out in Nature Communications! www.nature.com/articles/s41... It includes a thorough analysis and annotation of the single-cell transcriptomic atlas of an amphioxus neurula.
- Reposted by Sean MontgomeryPaper version of this really nice study is out now: www.nature.com/articles/s41... #epigenetics
- Reposted by Sean MontgomeryThe landscape of transcription factor promoter activity during vegetative development in Marchantia. Reporter fusions for most TF promoters, analysed expression patterns in Marchantia gemmae & build a map of expression domains to find key TFs in the stem cell zone academic.oup.com/plcell/advan...
- Reposted by Sean Montgomery[Please RT‼️] Mendel Early Career Symposium registration is open! Great speakers, small meeting, affordable & in Vienna. Great opportunity for #ECRs to network and get feedback. Please spread the word!!! #Mendel_ECR_24
- Reposted by Sean MontgomeryReally awesome paper by the Hannon lab showing dual H3K27me3/K9me3 at transposons and linking it to piRNA production! Cool parallels with work from Sandra Duharcourts Lab on paramecium!
- Reposted by Sean MontgomeryFirst major work fully made in the laboratory is out in @biorxiv-genomic.bsky.social: www.biorxiv.org/content/10.1.... DNA methylation allows recurrent endogenization of giant viruses in a #protist 🦠 closely related to animals, an unprecedented example of virus and eukaryotic host genomic mixture.
- Looks like a great new method and reads very nicely! Also a fantastic way to preprint: all supplementary figures are present AND links to supplementary tables!!
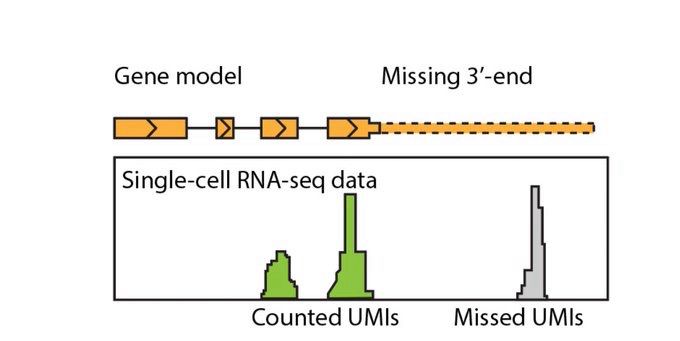
- Reposted by Sean MontgomeryCheck out our latest preprint! «GeneExt: a gene model extension tool for enhanced single-cell RNA-seq analysis» www.biorxiv.org/content/10.1... Presented in this thread by Grisha Zolotarov: twitter.com/zolotarg/sta...
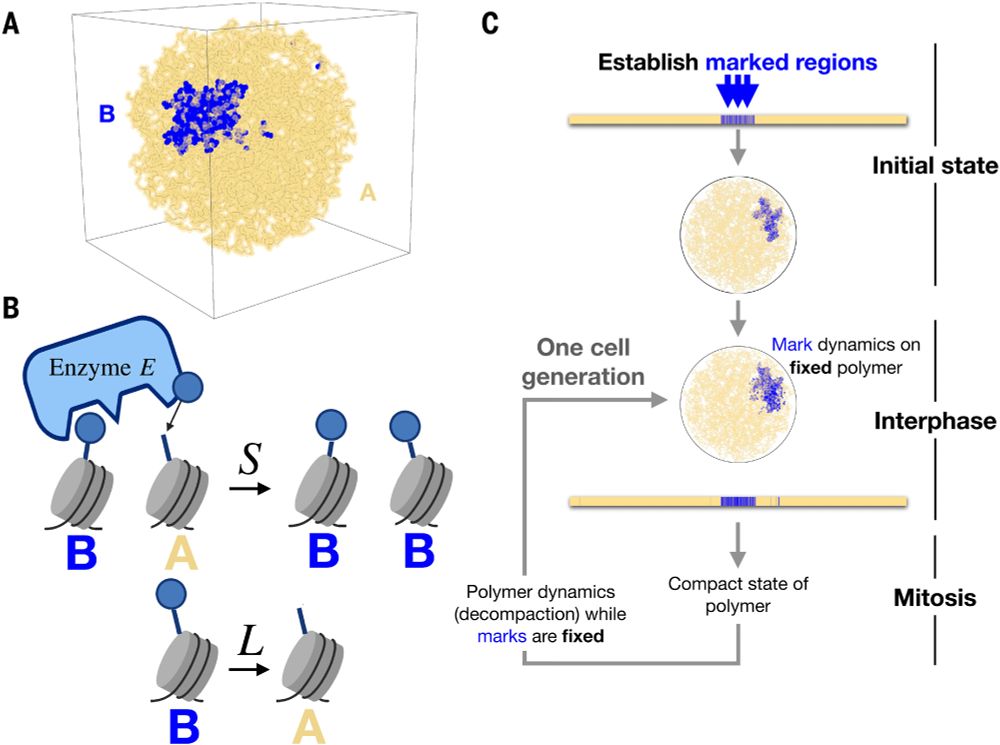
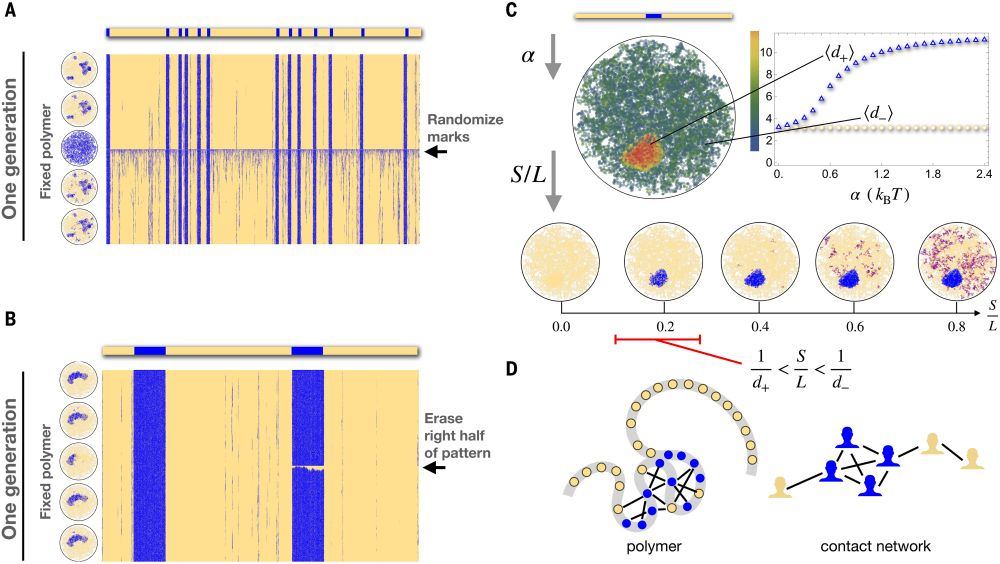
- Reposted by Sean MontgomeryPlease read our preprint: "G1 length dictates heterochromatin landscape" www.biorxiv.org/content/10.1...
- Reposted by Sean MontgomeryHow cell identity is preserved when cells divide? Owen, Osmanović, Mirny in a Science paper - Design principles of 3D epigenetic memory systems bit.ly/adg3053 - proposes that 3D genome folding is essential for passing down cellular memories MIT News bit.ly/adg3053mit also 👀 bit.ly/adg3053yt fr 16:35
- Reposted by Sean Montgomery🚨New Preprint🚨 In this study, we set out to determine how the de novo DNA methylation program impacts the 3D cis regulatory landscape. Comments welcome! This work was led by superteam Ana Monteagudo-Sanchez and @jrichardalbert.bsky.social www.biorxiv.org/content/10.1...
- Great to have this review out on what we know (so far) about imprinting in Marchantia! It was a good chance to summarize many of the musings we had on potential molecular mechanisms and biological implications! doi.org/10.1111/nph....
- Reposted by Sean MontgomerySometimes a paper stops you in your tracks. New work from Loppin’s group found that histone eviction during sperm development ensures that paternal chromosomes aren’t mistaken for maternal chromosomes in the Drosophila embryo. My Perspective: www.science.org/doi/full/10.... 1/2
- Reposted by Sean MontgomerySince I've introduced a few of my colleagues to this resource recently: www.phylopic.org has free to use silhouettes of a large number of organisms. These can be really useful for drawing out phylogenies (for the non biologists: basically a species-level family tree).
- Reposted by Sean MontgomeryHistones in bacteria (now exist even more, in their official, reviewer-sanctioned state): www.nature.com/articles/s41...
- Reposted by Sean Montgomery📢 Job alert! 👩🔬🌱🦠🧑🔬 Are you interested in #PlantEvolution, receptor kinases and/or immunity? Do you have a PhD in plant molecular biology? This position might be for you 👇 We are looking for a #postdoc to join our lab @ZMBP_Tuebingen Please RT! #PlantSciJob